|
NEWS
| 10/22/2025 | Hosted Dr. Rahmatullah Roche for a research talk at CCSB@PVAMU | Watch | |
| 10/17/2025 | Elected as a Faculty Advisory Council member for the College of Engineering at PVAMU | |
| 10/08/2025 | Presented our research works at CCSB@PVAMU | |
| 09/10/2025 | Awarded the RISE Undergraduate Research Grant by Research & Innovation Division at PVAMU | |
| 08/06/2025 | Attended Quantum Security and Machine Learning Workshop at the University of Tennessee at Chattanooga | |
| 07/28/2025 | Our EquiPNAS article is amongst the most-cited recent articles in Nucleic Acids Research | |
| 07/18/2025 | Served as a Data Science PFx Instructor at the Atlanta University Center Consortium | |
| 07/18/2025 | Attended the AI-assisted Cybersecurity Workshop at Tennessee Tech University | |
| 05/30/2025 | Contributed to the GET-PHIT Summer 2025 bootcamp | Snapshot | |
| 04/22/2025 | Awarded $5,000 in Google Cloud research resources | |
| 02/19/2025 | Presented our research works at CCSB@PVAMU | Watch | |
| 01/02/2025 | Our EquiRank paper is published in Computational and Structural Biotechnology Journal | |
| 12/18/2024 | Attended GenAI Faculty Summit organized by Google. Snapshot | |
| 10/12/2024 | Presented our EquiRank work at ICIBM 2024, held at Rice University | |
| 09/15/2024 | Awarded FEP award by the Roy G. Perry College of Engineering at PVAMU | |
| 08/31/2024 | Organized Machine Learning workshop jointly with Dr. Wang, sponsored by Google's TEC | |
| 08/23/2024 | Our EquiRank work has been accepted to ICIBM 2024 | |
| 07/19/2024 | Served as a Data Science PFx Instructor at the Atlanta University Center Consortium | |
| 04/09/2024 | Selected as 1 of the 20 Researchers for the Research Affinity Cohort (RAC) program of the National Data Science Alliance (NDSA) | |
| 04/05/2024 | Presented research work on Faculty Research Day at PVAMU R&I Week, 2024 | |
| 04/02/2024 | Graduate student Sheriff Adepoju presented a poster at PVAMU R&I Week, 2024 | |
| 03/12/2024 | Awarded the Google TEC Impact Fund, led jointly with Dr. Wang | |
| 01/28/2024 | Our EquiPNAS paper is published in NAR | |
| 01/02/2024 | I joined the Department of Computer Science at Prairie View A&M University as an Assistant Professor | |
| 12/05/2023 | Successfully defended Ph.D. dissertation. Acknowledgment | |
| 11/10/2023 | Accepted a tenure-track Assistant Professor position in the Computer Science department at Prairie View A&M University | |
| 10/25/2023 | Passed the CS@VT Research Defense | |
| 09/17/2023 | Our EquiPNAS preprint is available at bioRxiv | |
| 08/31/2023 | Our EquiPPIS paper has been accepted for publication in PLOS Computational Biology and available here | |
| 07/25/2023 | Awarded the Pratt Fellowship at VT | |
| 06/19/2023 | Served as a subreviewer | |
| 06/02/2023 | Our PIQLE paper has been accepted by Bioinformatics Advances and available here | |
| 05/09/2023 | Passed the CS@VT PhD preliminary exam | |
| 03/28/2023 | Our book chapter on homology modeling is published and available here | |
| 03/17/2023 | Our iQDeep paper has been accepted for publication in JMB | |
| 02/15/2023 | Our PIQLE paper is available at bioRxiv | |
| 12/17/2022 | Our EquiPPIS paper is available at bioRxiv | |
| 11/22/2022 | The CASP15 abstract has been published and is available here | |
| 11/16/2022 | Our book chapter on homology modeling has been accepted for publication to Springer Nature | |
| 8/19/2022 | Participated in the 15th Critical Assessment of Protein Structure Prediction experiments (CASP15) | |
| 07/31/2022 | Our iQDeep web server is released for beta testing | |
| 07/12/2022 | Our rrQNet paper is published in PROTEINS | |
| 06/20/2022 | Our DeepRefiner abstract has been accepted for the ACM-BCB 2022 Highlights Track | |
| 04/27/2022 | Won first place in Young Scientist Excellence Award competition at MCBIOS 2022 | |
| 04/08/2022 | Selected as one of the finalists for Young Scientist Excellence Award at MCBIOS 2022 | |
| 11/15/2021 | Passed the CS@VT PhD qualifier process | |
| 8/30/2021 | The CASP-COVID experiment paper is published in PROTEINS | |
| 8/11/2021 | Moved to Virginia. Snapshot | |
| 6/10/2021 | Served as a subreviewer at BIOKDD 2021 | |
| 5/17/2021 | Our DeepRefiner paper is published in Nucleic Acids Research | |
| 4/21/2021 | Our DeepRefiner paper is accepted by Nucleic Acids Research | |
| 4/21/2021 | Our review paper is accepted by Frontiers | |
| 12/20/2020 | Our DeepRefiner webserver is publicly released | |
| 7/16/2020 | Presented the poster for our aceepted QDeep method at ISMB 2020. Presentation | |
| 7/13/2020 | Our QDeep paper is published in ISMB 2020 special issue of Bioinformatics | |
| 6/12/2020 | Received ISMB 2020 fellowship award | |
| 6/6/2020 | Our QDeep paper is accepted by ISMB 2020 | |
| 5/18/2020 | Participated 14th Critical Assessment of Protein Structure Prediction challenge (CASP14) | |
| 3/13/2020 | Participated CASP_Commons (COVID-19, 2020) | |
| 5/30/2020 | Our method QDeep is accepted for a poster presentation at ISMB 2020 as part of the 3DSIG COSI | |
| 1/13/2020 | Our SPECS paper is accepted by PLOS ONE | |
| 5/1/2018 | Participated 13th Critical Assessment of Protein Structure Prediction challenge (CASP13) |
RESEARCH

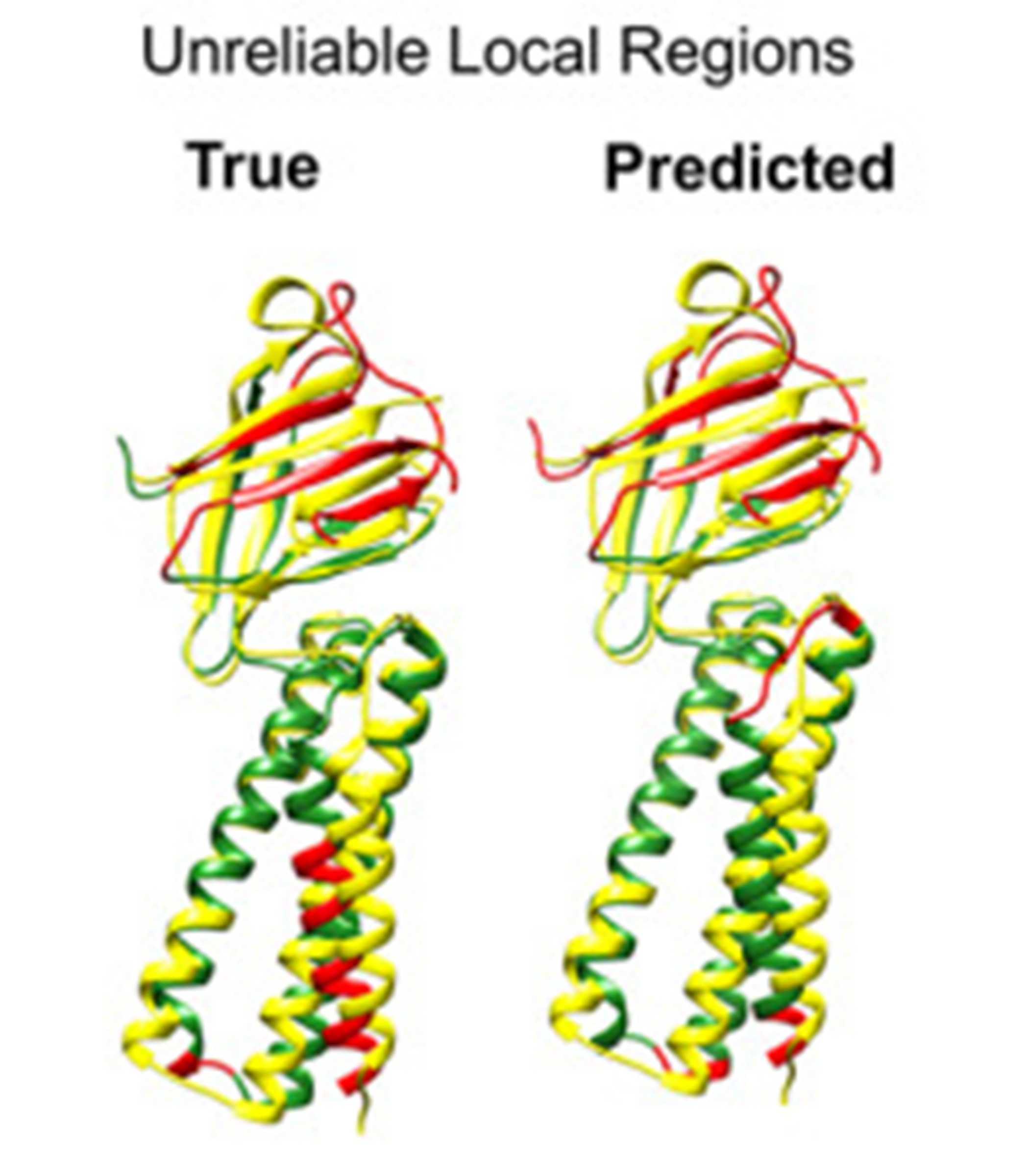
Protein Structure Quality Assessment
Deep learning for error estimation, structure scoring, and refinement
Read more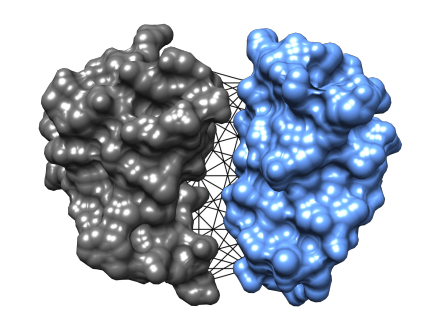
Protein–Protein & Protein–Nucleic Acid Interactions
Equivariant graph neural networks for binding site prediction, interface identification, and interface accuracy estimation
Read more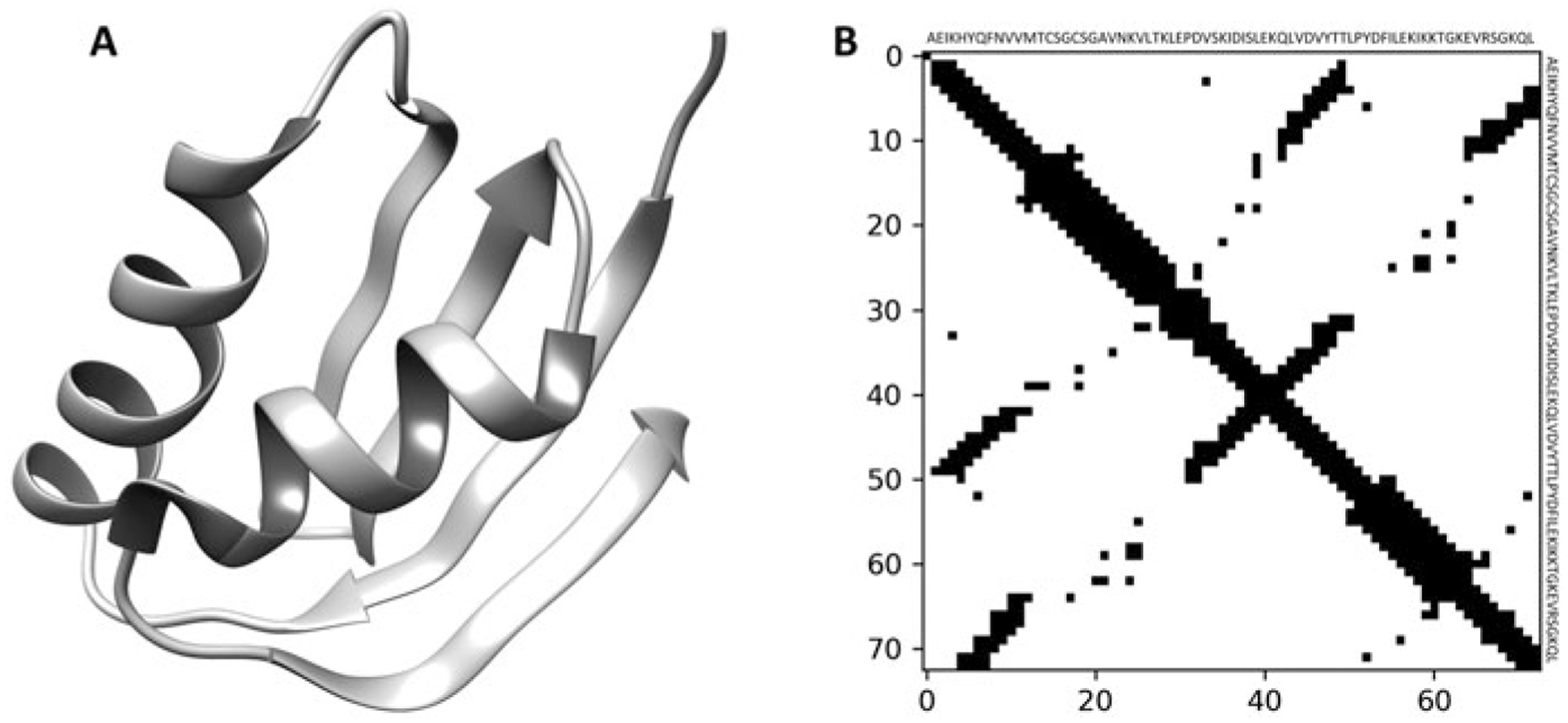
Protein Homology Modeling
Threading, template-based modeling, and contact-assisted threading for accurate protein structure prediction
Read more
Applied Machine Learning
AI applications in intelligent transportation and sustainable materials discovery
Read morePUBLICATIONS
-
2025
- Md Hossain Shuvo, Debswapna Bhattacharya.
EquiRank: improved protein-protein interface quality estimation using protein-language-model-informed equivariant graph neural networks
International Conference on Intelligent Biology and Medicine, 2024 Computational and Structural Biotechnology Journal, Volume 27, 160 - 170, 2025
PDF | GitHub
-
2024
- Rahmatullah Roche, Bernard Moussad, Md Hossain Shuvo, Sumit Tarafder, Debswapna Bhattacharya.
EquiPNAS: improved protein–nucleic acid binding site prediction using protein-language-model-informed equivariant deep graph neural networks
Nucleic Acids Research, gkae039, 2024 PDF | GitHub
-
2023
- Md Hossain Shuvo, Mohimenul Karim, Rahmatullah Roche, Debswapna Bhattacharya.
PIQLE: protein-protein interface quality estimation by deep graph learning of multimeric interaction geometries
Bioinformatics Advances, vbad070, 2023 PDF | GitHub - Md Hossain Shuvo, Mohimenul Karim, Debswapna Bhattacharya.
iQDeep: an integrated web server for protein scoring using multiscale deep learning models
Journal of Molecular Biology, 168057, 2023 PDF | Server - Rahmatullah Roche, Bernard Moussad, Md Hossain Shuvo, Debswapna Bhattacharya.
E(3) equivariant graph neural networks for robust and accurate protein–protein interaction site prediction
PLOS Computational Biology, e1011435 PDF | GitHub - Sutanu Bhattacharya, Rahmatullah Roche, Md Hossain Shuvo, Bernard Moussad, Debswapna Bhattacharya.
Contact-Assisted Threading in Low-Homology Protein Modeling
Methods in Molecular Biology book series, vol. 2627, 2023 PDF
-
2022
- Rahmatullah Roche, Sutanu Bhattacharya, Md Hossain Shuvo, Debswapna Bhattacharya.
rrQNet: Protein contact map quality estimation by deep evolutionary reconciliation
Proteins: Structure, Function, and Bioinformatics, (2022). PDF | GitHub
-
2021
- Andriy Kryshtafovych,..., Md Hossain Shuvo., ...,
Modeling SARS-CoV2 proteins in the CASP-commons experiment
Proteins: Structure, Function, and Bioinformatics, (2021). PDF - Md Hossain Shuvo, Muhammad Gulfam, Debswapna Bhattacharya.
DeepRefiner: high-accuracy protein structure refinement by deep network calibration
Nucleic Acids Research, Web Server Issue, gkab361 (2021). Selected for Highlight Talk at 13th ACM-BCB conference PDF | Server - Sutanu Bhattacharya, Rahmatullah Roche, Md Hossain Shuvo, Debswapna Bhattacharya.
Recent Advances in Protein Homology Detection Propelled by Inter-Residue Interaction Map Threading
of protein structural models
Frontiers in Molecular Biosciences, 8: 643752 (2021). PDF | In memoriam of Cyrus Chothia
-
2020
- Md Hossain Shuvo, Sutanu Bhattacharya, Debswapna Bhattacharya.QDeep: distance-based protein model
quality estimation by residue-level ensemble error classifications using stacked deep residual
neural networks
ISMB Proceedings (2020), Bioinformatics, 36(S1): i285-i291 (2020). PDF | GitHub - Rahul Alapati, Md Hossain Shuvo, Debswapna Bhattacharya. SPECS: Integration
of side-chain orientation and global distance-based measures for improved evaluation
of protein structural models
PLOS ONE, 15(2): e0228245 (2020). PDF | GitHub | Server
ACADEMICS
-
EDUCATION
- Ph.D. in Computer Science Virginia Tech Dec 2023
- M.Sc. in Computer Science Alabama A&M University July 2017
- B.Sc. in Computer Science and Engineering Bangladesh University of Business and Technology July 2014
-
AWARDS & HONORS
- Awarded FEP award by the Roy G. Perry College of Engineering at PVAMU September 15, 2024
- Awarded Google TEC Equity Collective Fund March 12, 2024
- Awarded the Pratt Fellowship at Virginia Tech July 25, 2023
- DeepRefiner has been accepted for the ACM-BCB 2022 Highlights Track August 7-10, 2022
- Awarded 1st place prize in Young Scientist Excellence Award competition at 18th annual MCBIOS Conference, 2022 April 27, 2022
- Awarded conference fellowship for MCBIOS 2022 April 25-27, 2022
- Awarded conference fellowship for ISMB 2020 July 13-16, 2020
- Awarded 2nd place prize at AAMU STEM Day-2016 April 14, 2016
- Awarded travel fellowship for IEEE SoutheastCon 2016 March 30 - April 3, 2016
-
POSTERS AND PRESENTATIONS
- M. H. Shuvo, S. Bhattacharya, D. Bhattacharya, "QDeep: distance-based protein model quality estimation by residue-level ensemble error classifications using stacked deep residual neural networks", ISMB 2020 Slide | Presentation
-
ACADEMIC EXPERIENCES
-
Assistant Professor
Prairie View A&M University
Jan 2024 - Present
1. COMP 3305: Analysis of Algorithms
2. COMP 3322: Software Engineering
3. COMP 1337: Computer Science II
4. COMP 1122: Computer Science Lab II
Show details -
Graduate Research Assistant
Virginia Tech
Aug 2021 - Dec 2023
Graduate Research Assistant:
Under the supervision of Dr. Debswapna Bhattacharya
VTech: Aug 2021 - Dec 2023
AU: Jan 2018 - Jul 2021 Show details - Graduate Teaching and Research Assistant Auburn University Jan 2018 - Jul 2021 Graduate Research Assistant: Under the supervision of Dr. Debswapna Bhattacharya (Jan 2018 - Jul 2021) Graduate Teaching Assistant: >COMP1210: Fundamentals of Computing I: Fall 2018, Spring 2019, Fall 2019, Spring 2020 >COMP 5970/6970: Computational Biology Show details
- Graduate Assistant Alabama Agricultural and Mechanical University Aug 2015 - Jul 2017
- Lecturer, Dept. of Stat, Math & Com Dhaka Commerce College Jan 2015 - Aug 2015
- Lecturer, Dept. of ICT Dhaka Cambrian College Aug 2014 - Dec 2014
 Office: 309 S.R. Collins Engr. Bldg.
Phone: (936) 261-9878
Email:
Office: 309 S.R. Collins Engr. Bldg.
Phone: (936) 261-9878
Email:  Google Scholar
Google Scholar